Pharmacogenetics
Challenges in Pharmacogenetic (PGx) Testing
Genetic factors account for 20-40% of the differences in individual drug metabolism and response1.US FDA reports costs for Adverse drug reaction (ADR) to be $136 billion annually2. Many technologies have inherent inefficiencies which drive up testing costs, waste resources and make PGx testing difficult and in impacts turnaround time. Typical issues are
- DUPLICATE SAMPLES – High assay failure rates force labs to run multiple replicates for each sample to avoid re-testing
- EXPENSIVE COST PER TEST – Even before unexpected inefficiencies, high reagent costs make testing expensive
- POOR REAGENT UTILIZATION – Running with less than a full run’s worth of samples wastes reagents and money
- MULTIPLE WORKFLOWS – Genotyping and copy number detection requires separate workflows
Ventola CL. Role of pharmacogenomic biomarkers in predicting and improving drug response: part 1: the clinical significance of pharmacogenetic variants. P T. 2013;38(9):545–560.
Centers for Medicare and Medicaid Services. Available from: https://www.cms.gov/Research-Statistics-Data-and-Systems/Statistics-Trends-and-Reports/NationalHealthExpendData/Downloads/proj2014.pdf. CMS; 2013. Accessed November 27, 2015.
PGx Solutions on MassARRAY
- Accuracy – Accurate detect genotype and copy number
- Combined detection – Simultaneously perform PGx copy number detection and genotyping using the same workflow
- Common workflow – Perform multiple panels during the same run using the same instrument
- Flexible Throughput – Process up to 100s or 1000s of samples per day with no minimum run size
- Efficiency – Obtain results in a single day with limited hands-on time
- Results Visualization – Automated software provides diplotype, halotype and CNV call in a single report
- Robust Performance – High assay success rate. No need for duplicate samples
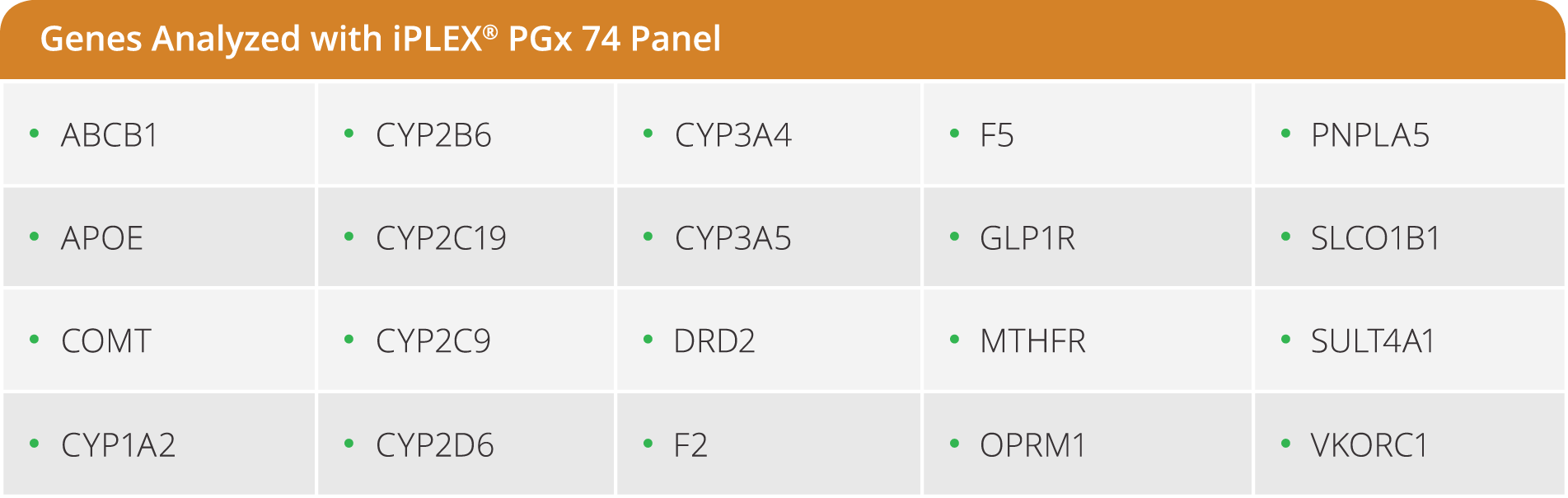
iPLEX Pro PGx74 Panel
The iPLEX Pro PGx74 panel is a pre-designed panel that targets the most relevant variants in 20 principle genes implicated in drug metabolism pathways. The panel provides genotype information for 69 SNPs/INDELs across 20 genes, plus 5 CNV targets in CYP2D6.
- Detects 74 targets across 21 of the most relevant genes
- Requires only 30 ng of DNA input
- from DNA to data in a single workday
- Test 32 samples in a single run
VeriDose™ CYP2D6 CNV Panel
CYP2D6 copy number variation (CNV) detection is a critical aspect of PGx testing as variations in the CYP2D6 gene affect drug metabolism. However, not all CYP2D6 alleles are functionally similar. Depending on ethnicity, up to 45% of the population possesses non-functional CYP2D6 “hybrid alleles” including *36, *13 and *68. Many copy number detection methods cannot differentiate between these non-functional “hybrid alleles” and other CYP2D6 alleles; resulting in an incorrect gene copy number or incorrect drug metabolism rate determination.
The VeriDose™ CYP2D6 CNV Panel accurately detects CNVs even in the presence of difficult to detect hybrid alleles. This assay can be run simultaneously with Agena’s genotyping panels. Seamless integration consolidates genotyping and CNV workflows.
- Broad Coverage – The VeriDose CYP2D6 CNV Panel is designed to interrogate 6 regions of the CYP2D6 gene in a single assay for broad gene coverage that eliminates the need for multiple assays or reflex testing.
- Hybrid Allele Detection – Accurate CNV calling; even in the presence of difficult to detect hybrid alleles.
- Requires only 10 ng of DNA input
- from DNA to data in a single workday
- Test 96 samples in a single run
Additional Pre-Designed PGx Panels
- CYP2C19: 31 SNPs in the CYP2C19 gene
- CYP2C9/VKORC1: 51 SNPs across 4 genes
- UGT1A1: 4 repeats in the UGT1A1 gene
- SLC6A4: INDEL and 1 SNP detection in the SLC6A4 gene
- DYPD
All assays are for research use only, not for use in diagnostic procedures.
Need a unique solution? SEQ-IT can help with custom assay design service.
Contact: massarray@seq-it.de
MassARRAY
Kontaktdaten
SEQ-IT GmbH & Co. KG
Pfaffplatz 10
67655 Kaiserslautern / GERMANY
Tel.: +49 (0)631 / 31670 – 0
E-Mail: info@seq-it.de



